Melanie Mallory, MS, lead bioinformatics scientist at ARUP Laboratories, played a key role in the development and validation of ARUP’s Cytomegalovirus Drug Resistance by Next Generation Sequencing assay.
Next generation sequencing (NGS) provides a more sensitive and rigorous method for detecting drug- resistant cytomegalovirus (CMV) variants, according to a new journal article authored by experts at ARUP Laboratories.
The article, “Development and Validation of a Next-Generation Sequencing Assay With Open-Access Analysis Software for Detecting Resistance-Associated Mutations in CMV,” was published on November 21, 2023, in Journal of Clinical Microbiology.
The article and the accompanying commentary describe the assay validation and analyze how NGS performs when compared with the traditional method of identifying drug resistance, Sanger sequencing. NGS analyzes multiple genes simultaneously and can detect drug-resistant variants at populations as low as 10% of the total viral population, whereas Sanger sequencing can only detect drug-resistant variants at populations of 20% or more.
For patients, a more sensitive assay that can identify more drug-resistant variants and at a lower percentage of the viral population may mean they receive a more effective drug therapy earlier in their treatment course and have better outcomes.
“NGS enables assessment of multiple genes [drugs] simultaneously, with higher resolution than is possible with standard Sanger sequencing,” said Kim Hanson, MD, MHS, ARUP’s section chief of Clinical Microbiology, medical director of Mycology, and the article’s senior author. “This may allow for earlier detection of resistance, and in turn, earlier prescription of optimal therapy.”
A coauthor of the article, Melanie Mallory, MS, lead bioinformatics scientist at ARUP, has played a key role in the validation and development of the NGS assay.
“This assay stands out based on how much we’re sequencing and how thoroughly we’ve validated,” Mallory said. “We performed additional experiments for CMV to ensure a robust validation, such as testing plasmids designed with as many mutations as possible to challenge the assay.”
Although CMV is a common virus that typically doesn’t cause illness in healthy individuals, it can pose a serious risk to individuals who are immunocompromised, particularly organ transplant recipients after their operations.
“CMV is the major opportunistic virus affecting organ and stem cell transplant recipients; it remains an important cause of posttransplant morbidity,” said Chief Operations Officer Adam Barker, PhD, NGS Infectious Disease medical director at ARUP and another coauthor. “Several CMV variants are known to confer drug resistance, so it’s important for clinicians to understand the specific variants to establish the right drugs for treatment.”
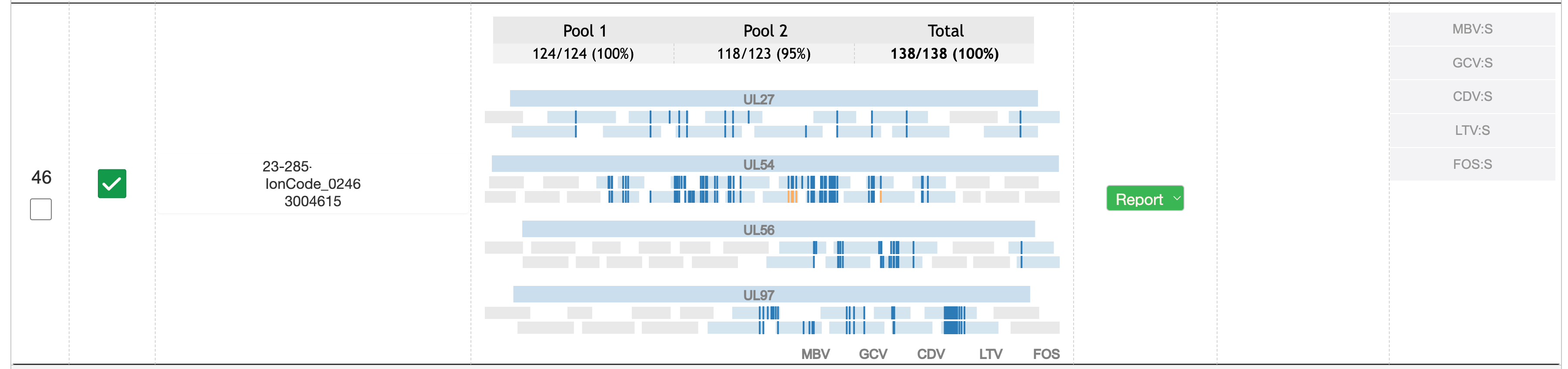
ARUP launched its CMV drug resistance assay by NGS at the end of 2022. The assay detects resistance mutations in UL27, UL54, UL56, and UL97 and reports susceptibilities and drug resistance for all five antiviral drugs currently available to treat CMV infection: cidofovir, foscarnet, ganciclovir, letermovir, and maribavir.
“Next generation sequencing enables us to provide significantly more information than traditional Sanger sequencing,” Mallory said. “By sequencing so exhaustively, we can reduce the risk for false negatives and more definitively identify the true drug-resistant variants with a single assay.”
According to Mallory, NGS provides about 10 kilobases of data compared with only 2 kilobases of data rendered by Sanger sequencing. Traditionally, subregions of two genes, U97 and UL54, have been sequenced by Sanger; however, the emergence of new drug-resistant variants and the availability of new drugs have created a need for the more extensive sequencing that NGS can provide.
During the validation, Mallory and the ARUP team found that NGS successfully identified resistant mutations that had been missed by the Sanger-based method in 16 samples. NGS can also more definitively identify variants when results are ambiguous or indeterminate with Sanger.
As part of the assay development, ARUP’s bioinformatics team developed an in-house software program to perform read alignment, call variants (ie, identify the variants), and interpret drug resistance.
“We have simplified the whole bioinformatics pipeline to provide a turnkey solution with usability features for the everyday lab technician,” said Keith Simmon, PhD, Research and Development scientific manager at ARUP and another article coauthor.
The software analyzes and calls variants on the same instrument that generates the sequence, which eliminates the need to transfer large volumes of data. Additionally, the software can analyze each run of multiple samples for potential errors, such as sample imbalance, and alert laboratory staff.
ARUP has made this software available to the public at no cost.
“Ideally, antiviral resistance testing should be performed near the patient for more rapid turnaround time to results,” Hanson said. “Providing software solutions for others could help facilitate in-house testing as well as promote CMV drug resistance research to advance the field.”
ARUP has also implemented a similar software program to aid in the assignment and reporting of drug-resistant variants in human immunodeficiency virus (HIV) and plans to continue to develop drug resistance detection assays for additional pathogens using NGS.
Kellie Carrigan, kellie.carrigan@aruplab.com