What Is Making My Patient Sick?

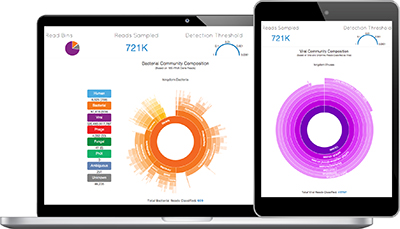
Dr. Robert Schlaberg co-developed Taxonomer, a new kind of pathogen identification tool capable of revealing all pathogens in a patient sample. The Bill & Melinda Gates Foundation awarded Dr. Schlaberg a $100,000 grant to use this technology to help decrease the high mortality rate of children with infectious diseases in resource-limited settings. Robert Schlaberg, MD, Dr Med, MPH, pulls up a colorful pie chart on his laptop. “This shows us a very high-level view of what microbes are present in this patient’s sample,” says Schlaberg, who specializes in molecular infectious disease testing at ARUP.
He clicks on a slice of the pie labeled “Viruses” and what look like purple tree rings appear, instantly revealing virus subcategories. Click. Now the “Bacteria” slice of the pie presents a set of orange tree rings.
Behind this interactive display is the processing work of Taxonomer, a new analysis software that sifts and sorts through millions of bits of information representing all known pathogens—those normally in our bodies and those that can make us sick— including viruses, bacteria, fungi, and parasites.
Taxonomer can identify an infection without the physician having to decide what to test for, something a PCR-based test cannot do. In other words, a doctor doesn’t have to suspect the cause of a patient’s infection, but can instead simply ask, “What does my patient have?” and Taxonomer will identify the pathogens. This means the patient is diagnosed more quickly, likely decreasing testing and hospitalization costs.
Taxonomer is an ultra-fast, metagenomic analysis software tool, meaning it can mine information from the vast amounts of genomic information extracted from microbial DNA. This DNA is found in the pathogens located in a patient specimen (i.e., blood, saliva). The amount of data that Taxonomer is extracting information from is immense; it must sift through millions of DNA sequences (fragments) to hunt for any known pathogen DNA.
Pioneering Technology Advances Diagnosis of Infectious Diseases
To understand the enormity of this information, imagine finding the novel Moby Dick shredded, then being given the task of locating the third letter in the 10th word, in the 23rd paragraph, in the 15th chapter. Now, find the equivalent letter in every other chapter. Now, patch together each letter and see if it forms a word—an identifiable known pathogen.
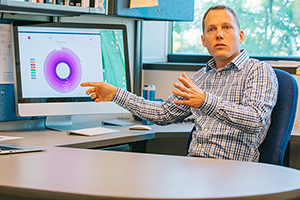
“While light microscopes allowed us to see what constitutes the blood and eventually what causes infections, Taxonomer is like a genomic microscope, allowing us a very detailed view of the processes going on in infected tissue,” says Schlaberg. The software’s analysis provides vital clues for detecting and treating infectious diseases.
Infectious diseases are one of the biggest killers in the world. Almost 2 million children under age 5 die each year from infectious diseases worldwide, yet many infections are treatable if the pathogen culprit can be quickly and accurately identified.
Take community-acquired pneumonia, for example. As one of the most common infections, it hospitalizes thousands annually, but no discernible responsible pathogen is identified in 20 percent of children and 60 percent of adults with pneumonia, according to findings published by Schlaberg’s collaborators in the New England Journal of Medicine last year.
This technology can be applied whenever we don’t know the cause of the disease, including when there are sudden outbreaks,” says medical epidemiologist Seema Jain, MD, with the Centers for Disease Control and Prevention. “We urgently need more accurate diagnostics to greatly enhance the ability of public health response and clinical care.”
“Not only can you interrogate a sample without knowing what you are looking for, but you can study the relationship of pathogens,” explains Andrew Pavia, chief of the Division of Pediatric Infectious Diseases at the University of Utah. “This can be helpful for hospital outbreaks in which the origin of infection might be an IV or specific drug. We’re able to fingerprint the organisms, which can help show that the pathogens came from a single source.”
“In the realm of infectious diseases, this type of technology could be as significant as sequencing the human genome,” says Taxonomer co-developer Mark Yandell, PhD, a professor of human genetics at the University of Utah and co-director of the USTAR Center for Genetic Discovery. “Very few people have inherited genetic disease. At some point, everyone gets sick from infections.”
The combination of speed, accuracy, and ease of use are Taxonomer’s stand-out characteristics against the backdrop of this rapidly evolving area of metagenomic technology. After a patient’s sample is sequenced, the data is uploaded via the internet to Taxonomer. In less than one minute, the tool displays a thumbnail inventory of all pathogens in the sample, including viruses, bacteria, and fungi.
“It’s tens to hundreds of times faster than similarly accurate tools,” adds Schlaberg. He points out that current diagnostic testing still relies heavily on growing pathogens in the laboratory, which may not provide accurate results and can be time consuming. While advanced PCR-based tests are much faster, they are only available for a limited number of pathogens. PCR stands for polymerase chain reaction, a method used to amplify sections of DNA for analysis.
Taxonomer can identify an infection without the physician having to decide what to test for. In other words, a doctor doesn’t have to suspect the cause of a patient’s infection, but can instead simply ask, ‘What does my patient have?’
Robert Schlaberg, MD, Dr Med, MPH Medical Director, ARUP Laboratories
While similar “catch-all” tests have been used in the past to study infectious disease outbreaks, the data analysis was not suitable for use in a diagnostic laboratory. Analyzing millions of DNA sequences took days or weeks; results were often difficult to interpret or not sufficiently accurate.
Taxonomer technology is currently being applied to developing universal pathogen-detection tests at ARUP Laboratories and by the start-up IDbyDNA based in Silicon Valley and Salt Lake City; the first ARUP test using this technology will go live this year.
Genes Have Expressions Too
Another unique feature of Taxonomer is its ability to analyze the patient’s own genetic material, in this case mRNA that shows which genes are turned on, providing information on how or whether the patient’s body is reacting to an infection. “As a clinician, this gives you a better idea, when we identify a pathogen, whether it is really the cause of the disease,” says Carrie L. Byington, MD, professor of pediatrics of the University of Utah and co-director of the Center for Clinical and Translational Science. Finding an infectious pathogen does not always mean that it causes the patient’s symptoms. Looking at other clues in how the patient’s body is fighting back can help doctors pinpoint the cause of illness. Taxonomer allows doctors to home in on the culprit and begin with a targeted treatment.

“Seeing how a host [patient] reacts is extremely valuable; I believe this is a paradigm shift in how we diagnose people. It is why I wanted to be involved,” says Byington.
“This tool will also allow us to determine if the patient is responding to a bacterial or viral infection when we don’t find a pathogen or when we find multiple potential causes,” adds Byington, who believes the tool is especially valuable for treating children because they experience more lifethreatening infections early in life. Since treatments for virus and bacteria are very different (for example, antibiotics don’t work against viral infections), matching a patient’s sypmtoms to the pathogen that is likely causing them can make the difference between giving the right treatment and one that doesn’t work at all.
Schlaberg returns to the shades of color fanning out on his screen, explaining how Taxonomer distinguishes between the patient’s response and the invasive pathogens. The tool sorts sequences into “buckets,” ones that belong to the patient and those that don’t belong to the patient. Then they are further subgrouped into viral and bacterial sequences and other groups. “Once we have that initial sort done, we can further classify relevant sequences to find out which microorganisms are in a patient sample. It can tell us not only about the presence of pathogens but also their absence.”
Sometimes discovering the absence of a pathogen is critical information. Consider the case of a transplant patient whose body may appear to be fighting an infection. Should the doctor prescribe a high dose of antibiotics, which could put the patient at risk for other types of infection if “good” bacteria are killed off, or should the doctor refrain from antibiotics and risk the patient developing sepsis, which can be deadly? Taxonomer’s pathogendetection capabilities could reveal that there are actually no infection-causing pathogens present; the body sometimes mimics infection-like symptoms.
According to Schlaberg, the possibilities with Taxonomer extend beyond diagnostics. The tool has already uncovered a previously unknown virus from one patient sample. There’s likely much more where that came from, meaning Taxonomer could accelerate discovery of never-before documented causes of illness, including emerging infectious diseases set off by rapidly evolving pathogens that seed new outbreaks.

As a clinician, this gives you a better idea, when we identify a pathogen, whether it is really the cause of the disease … Seeing how a patient reacts is extremely valuable; I believe this is a paradigm shift in how we diagnose people. It is why I wanted to be involved.
Carrie L. Byington, MD, Co-Director, Center for Clinical and Translational Science, University of Utah
“We don’t know the diversity that exists within those pathogenic organisms, within a given species, but we’re generating that information very quickly now,” adds Schlaberg, pointing out that the genomes of viruses and bacteria are much smaller than the genome of humans, so it is more feasible and faster to generate and analyze this data.
Such information could set forth new diagnostic tests, preventions, and treatments. “The implications for discovery are what makes this so exciting,” says Yandell. “The potential is huge.”
Fighting Childhood Mortality
Dr. Schlaberg Receives Prestigious Gates Foundation Grant
In May, ARUP Medical Director Robert Schlaberg, MD, Dr Med, MPH, was one of 43 individuals globally to receive a prestigious grant from the Bill and Melinda Gates Foundation for a project to help decrease the high mortality rate of children with infectious diseases in resource-limited settings.
The $100,000 grant is part of the foundation’s Grand Challenges Explorations, which fosters innovation to solve global health and development problems. More than 1,400 applications were received.

Most of these deaths are preventable; it’s a tragedy. To prevent them, you first need to know what is causing the disease. With this technology, we have the methods to make a huge difference.
Robert Schlaberg, MD, Dr Med, MPH Medical Director, ARUP Laboratories
His proposal, titled Universal Pathogen Detection in Post Mortem Tissues, will use universal pathogen detection based on next-generation DNA sequencing to identify fatal infections. Using RNA sequencing and Taxonomer, a new analysis tool co-developed by Schlaberg, scientists will be able to identify the cause of death by detecting all known pathogens, including viruses, bacteria, fungi, and parasites, in post mortem tissues. Scientists will also differentiate infectious from noninfectious causes of death by immune profiling.
“The diagnostic tests that pick up on what is causing the child’s infection are often lacking, and children may not be seen by a healthcare provider in time,” says Schlaberg, who notes that some 2 million children under age 5 die each year from infectious diseases. Most of the fatalities stem from different types of pneumonia and diarrhea-causing infections, but hepatitis, meningitis/encephalitis, and sepsis are also culprits.
Schlaberg’s findings could ultimately help plan vaccine and prevention efforts and provide recommendations on treatments. “While we know the cause of many severe infections, such as malaria, in resource-limited settings, things become much more difficult when routine treatments for these common causes fail,” explains Schlaberg, pointing out that doctors may cast a wide net and base guesses on the most common known causes, which often happens with pneumonia. “Knowing the less common or unexpected causes will help design better preventative measures and treatment programs,” explains Schlaberg, who is also an assistant professor of clinical pathology at the University of Utah School of Medicine.















 HOME
HOME