ARUP’s next generation sequencing (NGS) tests are now faster and more precise thanks to the launch of Rio, a new world-class bioinformatics pipeline and analytics platform.
Rio amplifies ARUP’s existing cloud computing capabilities to move data more efficiently and with greater accuracy to speed targeted test results to clinicians for analysis and improved medical decision-making for patients.
“Rio’s flexible, scalable architecture provides a rock-solid foundation on which ARUP’s genomics future will be built,” said NGS Informatics Supervisor Brendan O’Fallon.
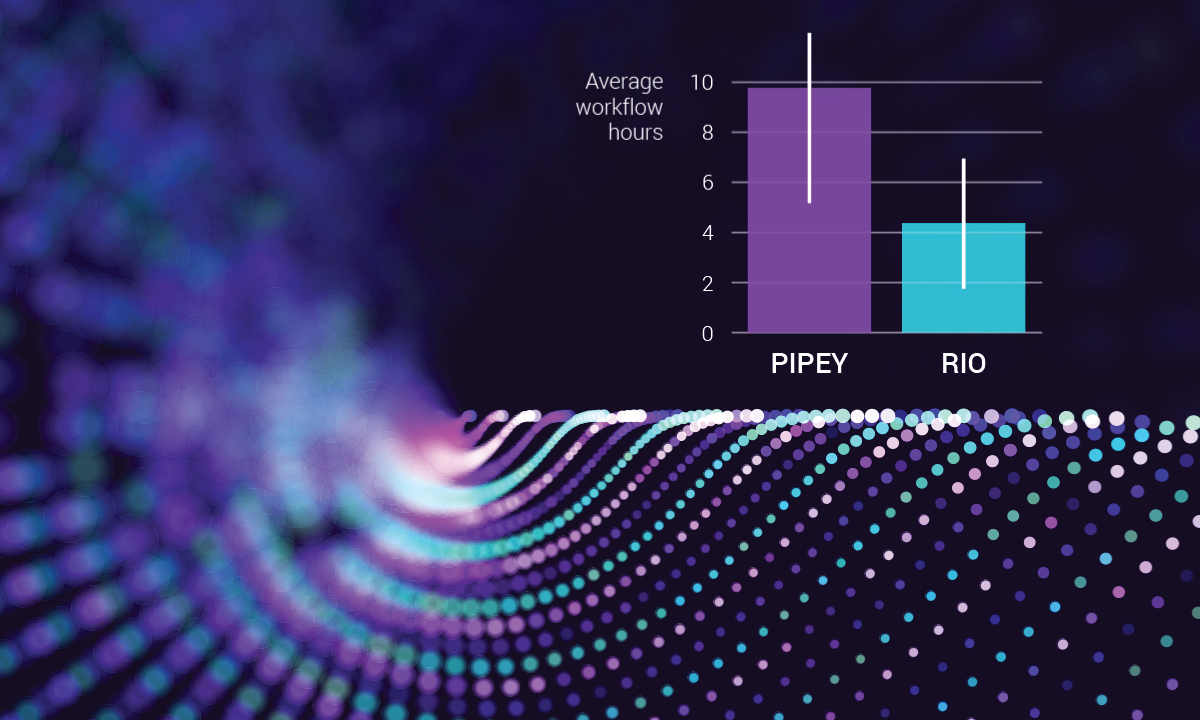
The platform became fully operational in 2021 and is living up to its promise. Lab data show that, with Rio, average test workflow times have been reduced from nearly 10 hours to about four.
Rio replaces ARUP’s first cloud computing pipeline, Pipey, which was introduced in 2018. The pipeline supports the work of medical directors and multiple labs, including Genomics and Oncology.
Like its predecessor, Rio was built in-house by a team of bioinformaticians, data engineers, software programmers, and system administrators over a period of about 18 months, said Sarah Broderick, manager of the Biocomputing Department. The Rio system was validated using the most recent recommendations from the College of American Pathologists (CAP).
Both the Rio and Pipey platforms were designed to improve the quality of the NGS pipeline by scaling testing analytics while processing large sample volumes and supporting rapid turnaround times. The latest version of Rio processes samples about two times as fast as Pipey, enabling shorter test turnaround times and lower processing costs. And perhaps more importantly, the Rio architecture makes it easy to incorporate new tools and features into production workflows.
The pipeline is a collection of standardized tasks that transform terabytes of raw data generated by sequencers from extracted specimens into lists of genetic variants and any of the associated annotations for interpretation by medical directors and PhD-level clinical variant scientists.
Moving to cloud computing in 2018 was an important step for ARUP. Tapping into the cloud increased ARUP’s data storage capacity and data management capability exponentially, while enhancing protections for patient information. More importantly, it allowed ARUP to process thousands of specimen samples in the same amount of time it previously took to process just one.
Pipey was pivotal in the transition to the cloud, but it also had its limitations. For example, the Pipey architecture analyzed every gene present in a sample, regardless of whether it was part of the ordered test, which made it much less efficient. Rio’s capabilities make the process faster because it only analyzes the test-specific genes. It’s just one example of how ARUP bioinformaticians have rewritten Pipey’s older, slower tools to increase the speed, in some cases, by a factor of 10 or more, O’Fallon said.
“It’s hard to overstate the importance of speed in this field when even one day can have a significant impact on the outcome for a patient in neonatal intensive care or a patient waiting to start cancer treatment,” said Hunter Best, PhD, scientific director of NGS and Biocomputing. “The updates to our bioinformatics pipeline introduced with Rio provide us with better means to give patients accurate answers in a quicker fashion.”
Jennifer Dobner, jennifer.dobner@aruplab.com